Alignment Systems
Introduction
VocBench 3 can use remote services to compute alignment between two datasets (associated with distinct projects), as described in the page on the alignment validation tool.
Alignment services API
The communication between VocBench 3 and a remote alignment service is carried on using a REST API, which supports activities including task management and settings retrieval. This API has been formally described using the OpenAPI Specification format. Such an API description can be used in a number of tools to perform several API-related activities, such as generation of server stubs, generation of client libraries, testing, documentation, etc. In particular, code generators (such as Swagger Codegen, with its online editor and OpenAPI Generator) can significantly help to develop a wrapper for an existing matching system, supporting dozens of programming languages and frameworks, while also fostering compliance with the API specification.
The description of the current version of the API is available online at this address http://art.uniroma2.it/maple/specs/alignment-services-2.0.0.yaml
See the section below for a reference of the API.
Alignment process
We assume that the user chose two align two datasets stored as projects in VocBench 3. Metadata about these datasets and other, potentially available resources, should be available in the metadata registry. The following description will be grounded in the alignment validation tool: in particular, the user is assumed to be at the point of creating a new task on a remote system.
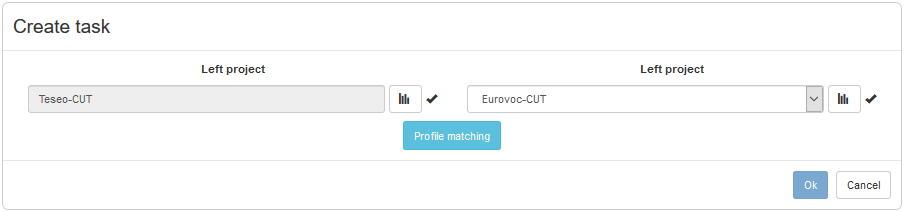
- When the user clicks the "Profile" button, VocBench 3 uses MAPLE to analyze the matching problem,
obtaining a report describing the alignment scenario.
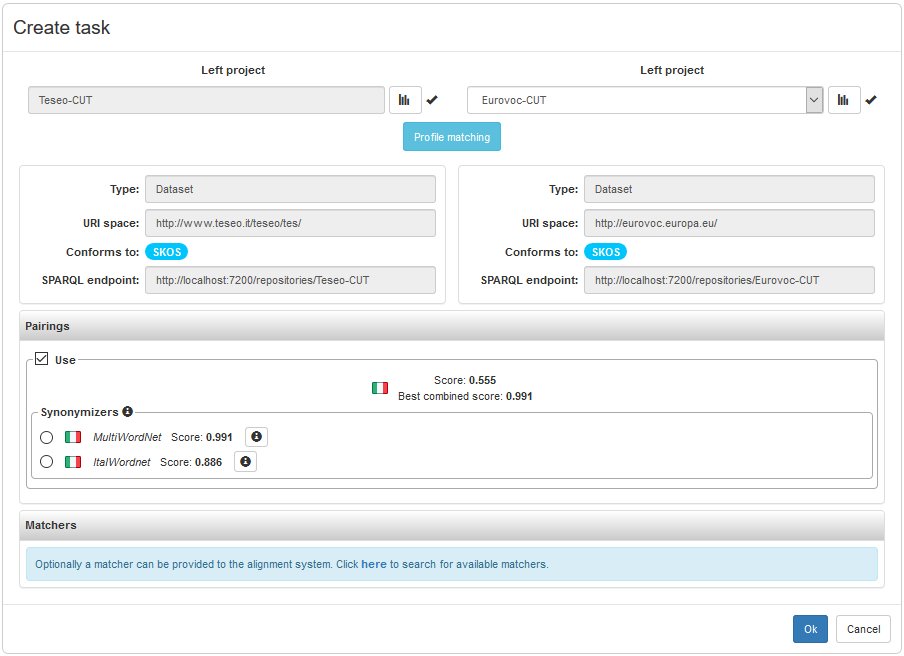
At the top of the scenario, we can find metadata about the input datasets, including their namespace (called URI space), knowledge model (labelled conformsTo) and SPARQL endpoint.
MAPLE is unaware about the actual algorithm used to match the input datasets, but it assumed that their lexical content plays a pivotal role. In the terminology of MAPLE, based on the metadata model LIME, a set of labels of a dataset is called a lexicalization set. Based on the previously stated assumption, an important part of the scenario description consists of a list of pairs of lexicalization sets for the input datasets: the pairings section in the figure. Each pairing is intended to suggest strategy to compare the input dataset at the lexical level, possibly benefiting from a wordnet-like resource.
-
the user can establish a concrete scenario definition, by choosing among the alternatives
proposed in the scenario. Examples of entries found in different scenarios are:
- lexicalization pairings (i.e. couple of lexicalizations, one from each of the datasets to be aligned)
- language resource (if any) to use as a support to the alignment (e.g. a WordNet in Italian could be used as a lexical bridge for augmenting the potential lexical overlap between two lexicalizations in Italian
- alignment chains: instead of relying on language, it is possible to use existing alignments to a third resource as a bridge
- The choice of the scenario is an optional step: as an alternative, the system will create a scenario definition by choosing just the first pairing (the one with the highest score), discarding any language resource.
-
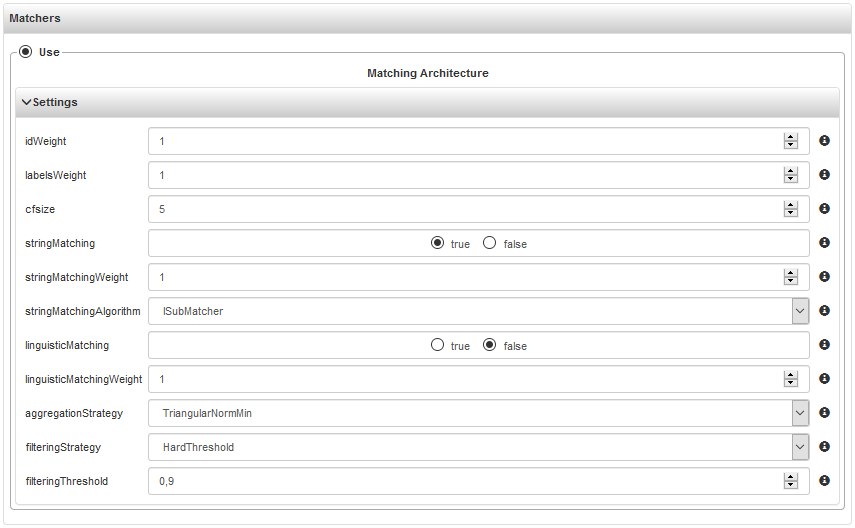
once a scenario definition has been established, the user can click the "search" link in the
"matchers" panel in the bottom, to retrieve matchers configurations suitable for this scenario
definition.
this step is optional, as the alignment service is expected to have a default configuration that is suitable for most situations. However, at user's willingness, this step allows to act on the configuration knobs exposed by alignment service. As they depend on the actual scenario definition, these matchers configuration shouldn't offer meaningless options: e.g. different options related to synonym expansion if no language resource has been selected (and the service doesn't embed any).
-
the user clicks on the button "OK" to create a new alignment task, based on the alignment plan,
consisting of:
- scenario definition
- optional settings
Compliant services
Hereafter, we report on alignment systems that have been made compatible with VocBench 3 by implementing our alignment API.
Ge.no.ma
Ge.no.ma is an Ontology Matching Environment that provides the user with a powerful tool to design and test ontology matching architectures.
Genoma is available, on its downloads page, as an archive called genoma-alignment-service-2.3.5.zip. To execute Genoma as a VocBench 3 service, it is sufficient to unpack the archive and execute either start.bat or start.sh, depending on whether the host operating system is Windows or a UNIX system.
Once launched, Genoma will be listening on address http://localhost:7575. Opening this address in the browser should load an interactive documentation (based on Swagger UI), as in the following picture.
NAISC
NAISC is an automated linking tool developed at the Insight Centre for Data Analytics. 'Naisc' means 'links' in Irish and is pronounced 'nashk'.
NAISC has its own REST API, which is being extended with further endpoints complying with our API specification. This support is currently available on the dev branch.
To launch NAISC as a VocBench 3 service, it is necessary to:
- download the dev branch
- on Windows, inside the folder naisc-rest:
- replace the file models (which is intended to be a symbolic link) with an empty directory
- replace the file configs (which is intended to be a symbolic link) with a copy of the homonym directory in the root of the projects sources
- execute either gradlew.bat (on Windows) or gradlew (on a unix-like system)
- armed with patience, wait until Naisc has downloaded its models
When running, Naisc listens for request at this address http://localhost:8080/naisc-rest/maple. Opening this address in the browser should return metadata about the service as a JSON object.
Alignment Bootstrap
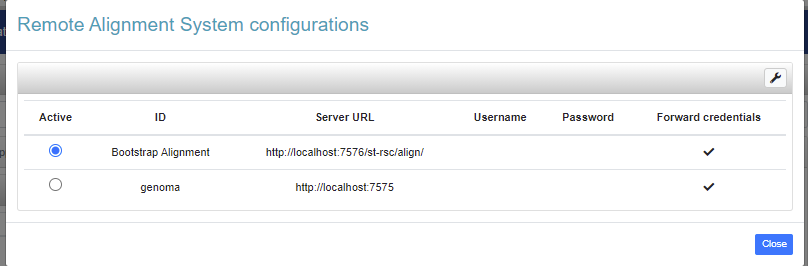
The Alignment Bootstrap is a set of services currently hosted on the ST Remote Service Compendium (ST-RSC), which exploits alignment chains (see previous section) to generate alignments between datasets bridged by alignments to a third one.
Once the ST-RSC service has been activated, the Alignment Bootstrap services will be listening on address http://localhost:7576/st-rsc/align (the standard url/port can be changed by modifying the startup script), so this is the URL to be used when connecting VB to the ST-RSC server.
ST-RSC's services are described in a YAML file, the model of which is based on OpenAPI 3.0.1. Note that the ST Remote Service Compendium offers other services, such as the Skos Diffing.